Our laboratories and research are directed towards understanding the mechanism by which eukaryotic organisms keep time on a daily basis, and how this capacity to keep time is used to regulate metabolism and development. Circadian clocks with fundamentally identical characteristics are found in all groups of eukaryotic organisms, but the uses to which these clock are put reflects the diversity of evolution. Phylogenetically this ranges from the control of cell division and enzyme activities in unicells, to a firmly established involvement in plant and animal photoperiodism and in avian and insect celestial navigation, to multiplicity of human systems including endocrine function, work-rest cycles and sleep, and drug tolerances and effectiveness. Cell division in many tissues and organs within the human body is regulated on a daily basis by the clock, providing the theoretical basis for chronotherapy of cancer, and manipulation of internal circadian rhythms provides treatments for several kinds of sleep and affective disorders. The general layout of the feedback loop that makes up the clock and the identity molecular identity of some of the components, particularly the heterodimeric positive elements WC-1 and WC-2 that serve as positive elements in the loop, appear to be conserved among the Crown Eukaryotes (Dunlap Science 280, 1548-49, 1998; Cell 96, 271-290, 1999).
We are pursuing the analysis of the clock chiefly by using as a model system the fungus Neurospora crassa, although similarities between this basic model system and the mammalian circadian oscillator have led us to examine the molecular biology of mammalian clocks also. As a lower eukaryote, Neurospora has a biological clock that is functionally equivalent to those seen in higher organisms, but at the same time Neurospora is a simple organism upon which can be brought to bear all of the current techniques of classical and molecular genetics and biochemistry.

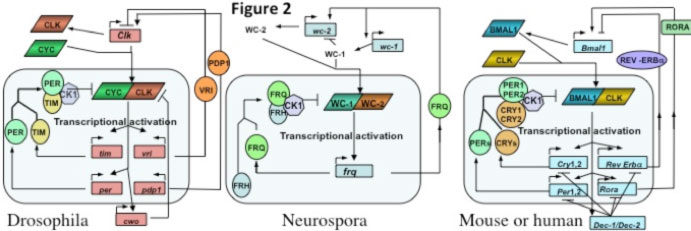
Circadian biological clocks represent a common overarching aspect of cellular and organismal regulation, yet all clock feedback loops close within single cells. Fig. 1 shows cartoon for eukaryotic clocks in the fungal/ animal lineage, a transcription-translation based feedback loop where transcription factors as positive elements drive expression of genes encoding proteins that block the activity of the negative elements.
Clocks in fungi and animals share a common regulatory architecture. "Positive Elements" are heterodimeric transcription factors, two proteins that interact via PAS domains (e.g. WC-1 &WC-2 in Neurospora, BMAL1 &CLOCK in vertebrates) and that share extended sequence similarity as well as common functions. In all cases, circadian output derives in part from the action of the clock-regulated positive elements on expression of clock-controlled genes (so called ccgs) whose products in general do not feed back onto the central clock loop.
Overview of the circadian oscillator mechanism
Cycles employing virtually identical regulatory architecture occur in fungi and animals. Circadian feedback loops from the fungal/animal lineage in figure 2 serve to highlight similarities and differences. The shaded rounded rectangle at the bottom of each encloses the "core feedback loop", those activities are generally essential for the clock, and the rest of the figures describe coupled feedback loops that provide stability and robustness, or that link to output. For instance, in the Neurospora core loop a WC-1/WC-2 complex drives expression of frq (Froehlich et al., 2002; Froehlich et al., 2003), and FRQ interacts with FRH to make the negative element leading to WC-1/WC-2 inhibition (reviewed in Baker et al., 2011; Dunlap, 2006, 2008; Dunlap and Loros, 2006; Dunlap et al., 2007). In mammals the CLOCK/BMAL1 drives expression of Per1, Per2, Cry1, Cry2 to form a negative complex; in Drosophila the heterodimer is CLK and CYC and the negative elements PER and TIM. Additional proteins join the negative element complexes that apparently act through recruitment of CK1, and other kinases, and modifiers, to depress the activity of the transcriptional activators (reviewed in Baker and Dunlap, 2010; Belden and Dunlap, 2008; Mehra et al., 2009a). The same basic clock operates in nearly all cells in these organisms and controls 5-20% of gene expression depending on organism/cell type. Ccgs (output genes) are regulated via a transcription factor hierarchy (reviewed in Chen et al., 2010b; Chen et al., 2009; Loros et al., 2007).
All three feedback loops use heterodimeric transcription factors interacting via PAS domains to drive expression of protein complexes that feed back to reduce the activity of the heterodimer (see table adapted from Baker et al., 2011). In Neurospora this occurs via FRQ-promoted phosphorylation of the WC-1/WC-2 heterodimer (reviewed in Dunlap et al., 2007, Mehra et al., 2009a; Baker et al., 2011), and this phosphorylation-mediated activity reduction almost certainly also happens in the fly and mammalian clock although the mechanistic link has yet to be established. Sequence similarity between fly and mammalian PER and the positive elements (CLK & CLOCK, CYC & BMAL1) extends to weaker but still significant similarity between BMAL1, CYC, and WC-1 (Lee et al., 2000). VRI (in flies) and REV-ERBα (mammals) act in a coupled loop to reduce expression of a positive element, and RORA, PDP1 and FRQ promote expression of one of their activators. In the fly brain clock CRY is a photoreceptor with no role in the feedback loop, but mammals use CRYs as the major repressor; consensus has neither
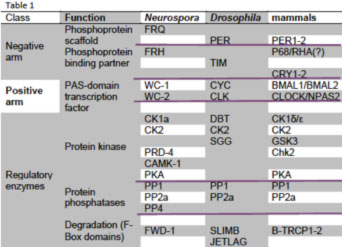
mPER3 nor mTIM in the mammalian core loop. There are swaps in DNA binding domains too: the nuclear receptor REV-ERBα acts through a RORE, but in flies VRI is a bZIP transcription factor binding a V/P-box; WC-1 and WC-2 use Zn fingers instead of bHLH domains as do their mammalian counterparts. Light resets the fungal and mammalian clocks the same way, via induction of negative elements (FRQ or mPER1, Crosthwaite et al., 1995; Shigeyoshi et al., 1997) whereas flies reset through CRY-mediated turnover of TIM. Kinase and phosphatase (e.g. CK1, CK2, CHK2, PKA, PP2A) functions are conserved. Two homologs in mammals perform many functions done by one protein in flies (i.e. two PERs, CLOCK and NPAS2, etc.). Aficionados will recognize that additional proteins have been associated with both activating and repressing complexes, especially histone-modifying proteins both in Neurospora and in mammals. These are generally helpful but not essential proteins.
Overview of the means of output from the circadian oscillator
A basic clock with similar regulatory architecture operates in nearly all cells in the human body and is also found in cells of model systems like Drosophila or Neurospora. In people the SCN clock coordinates rhythms by dictating sleep wake and feeding times and thereby directly or indirectly controlling cellular clocks; estimates based on organ-specific clock abrogation suggest that only about 15% of rhythmic genes are directly controlled by systemic signals; most of the ccgs, about 85%, are controlled by clocks operating at the cellular level. These tissue-specific genes are the workhorses of circadian regulation, their circadian biology driving the rhythmic physiology of the tissues. How does this work?
The major part of circadian output derives from clock regulation of transcription of genes, the ccgs making products that do not impact the feedback loop itself. Work from our lab pioneered the identification of ccgs (Loros et al., 1989; reviewed in Loros et al., 2007). Global circadian regulation of transcription is thought to occur chiefly through a hierarchy of factors, in the same way as light regulation in fungi (Chen et al., 2010b; Chen et al., 2009a,b). Lastly there are physiological cycles ("FLOs") that can free run on their own or can be coupled via unknown means to the circadian metabolism of the cell. Although poorly studied in mammalian cells, these are well known in Neurospora, perhaps because it is among the most intensively studied cellular circadian systems. A major effort in the last four years went toward demonstrating that such FLOs are only conditionally connected to the circadian core and can run on their own, even controlling aspects of metabolism, while not impacting the core clock (e.g. Loros et al., 2007; Mathur et al., 2011; Shi et al., 2007).
Similar pathways probably work in mammals although only the transcriptional network has received much attention. Clock/BNMAL1 (C/B) is active driving gene expression in the late afternoon, and targets include some transcription factors (TFs) (e.g. DBP, TEF, HLF, KLF10) that in turn regulate their own banks of genes phased a few hours after the peak of direct C/B targets. However a truly useful clock must drive expression of genes peaking at all times of day, and this comprises much of the rest of true circadian output. C/B also drives strong circadian expression of REV-ERB which represses BMAL1 expression via RORE elements, cis-acting elements that are also bound by the transcriptional activators ROR αβγ. These proteins elicit a rhythm in BMAL1 expression that reinforces overall rhythmicity although, as with WC-1, it is dispensable for rhythmicity per se. A major significance of the REV-ERB and ROR accessory loop, however, is in the other genes they regulate: these include direct RORE-regulated targets (e.g. the transcriptional co-regulator RIP140, about which you will read much more later) that peak in the late night in some cells. Among the targets regulated by REV-ERBs and RORs is the TF E4bp4, and genes regulated by E4bp4 will be induced a few hours later in the early morning. In the aggregate, between genes directly regulated by C/B in the late afternoon, by the PAR bZIP and KLF10 TFs regulated by C/B that are phased a few hours later in the evening, by the interplay of REV-ERBs and RORs driving expression in the night, and by E4bp4 phased a few hours later, expression spanning most of the day can be achieved. Combinations of different cis-acting elements can further modulate the activity of these secondary TFs, and cell-type-specific posttranslational modifications (PTMs) can alter phase in some cells so all circadian phases are accessible.
Neurospora remains a versatile system for understanding clock-regulated gene expression, and many paradigms originating there have been readily applied to mammals. Moreover, comparisons of mouse, rat, rhesus macaque, and human transcript arrays show that the antiphasic relationship between E-box and RRE cis-elements is conserved, so insights from mice are very likely to inform human circadian physiology. Given this, a major question for circadian biology is now how these factors are coordinated with physiology at the cellular, tissue, and organ system level to appropriately modulate metabolism.
All the feedback loops in cells from the fugal/animal lineage are characterized by daily synthesis and turnover processes that should not in themselves require 24 hours. Thus, the cutting edge questions remain (1) what is the nature of the time lags; (2) what are the essential core components, and which ones are helpful; (3) what is the mechanism through which they act; (4) how do their actions account for canonical clock properties. Because resetting is now largely understood, (4) is now focused on temperature compensation.
Overall then we are interested in understanding how the circadian system in organized in Neurospora and in mammals, and why evolution has done it in this way. For each of our approaches, our aim is to understand the organization of cells as a function of time, and ultimately to achieve an appreciation of life as a four-dimensional process. There is every reason to believe that the circadian system of Neurospora will be the first circadian system to be completely described at the molecular level, and that what we learn from both fungi and mice will lead us to a better appreciation and understanding of circadian timing in people.