 How do proteins in a human cell interact with one another?
How do proteins in a human cell interact with one another?
How do these protein interactions result in biological activity and disease?
How can we control this behavior with smaller molecules?
These three broad and interrelated questions are those our research group tries to answer, specifically as applied to a selection of proteins involved in cellular signaling pathways and enzymatic transformations. The tools we use to address these questions are diverse and reflect the multidisciplinary nature of our laboratory. These include organic and peptide synthesis, biochemistry, molecular biology, and biophysics (with the occasional excursion into structural biology). Operationally, we apply these to first investigate the properties of protein-ligand and protein-protein interactions at the fundamental level. The next stage then uses what we have learned to develop compounds with explicit utility, either as cellular probes or lead agents for drug discovery. At present, our projects grouped within one of two categories: (I) the PDZ domain family of proteins; and (II) bioactive small molecule organic compounds. What follows is a very minimalistic synopsis of our research activities within these two spheres.
I. PDZ Domains and the Inhibition Protein-Protein Interactions
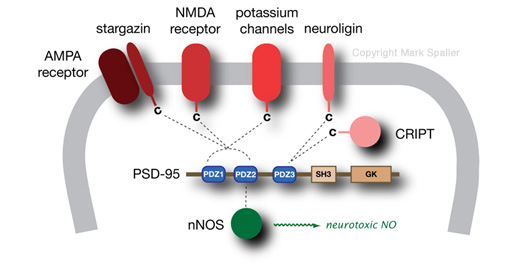 The PDZ domain is a structural motif found in hundreds of mammalian proteins. These operate within different physiological contexts, but many of primary interest are found inside the brain, and affect many critical neuronal functions. These include scaffolding proteins that serve to cluster and localize cellular membrane receptors and facilitate signal transduction. A significant part of our work to date has focused on postsynaptic density-95 (PSD-95), a prominant PDZ domain-containing protein that functions within networks that also involve NMDA and AMPA receptors. With three distinct PDZ domains (PDZ1, PDZ2, PDZ3), PSD-95 is a hub for a variety of transient protein-protein interactions. One particularly important role of PSD-95 is as a regulatory connection to neuronal nitric oxide synthase (nNOS). nNOS has been implicated in several neurotoxicity behaviors, prime among these being a result of brain ischemia and stroke. Molecular intervention at the level of PSD-95—namely, to create selective cell-permeable inhibitors that could target each PDZ domain—would provide for both useful research tools to further investigate this pathway. In addition, these ligands could constitute potential therapeutic agents for treating disorders resulting from faulty or unregulated nNOS activity.
The PDZ domain is a structural motif found in hundreds of mammalian proteins. These operate within different physiological contexts, but many of primary interest are found inside the brain, and affect many critical neuronal functions. These include scaffolding proteins that serve to cluster and localize cellular membrane receptors and facilitate signal transduction. A significant part of our work to date has focused on postsynaptic density-95 (PSD-95), a prominant PDZ domain-containing protein that functions within networks that also involve NMDA and AMPA receptors. With three distinct PDZ domains (PDZ1, PDZ2, PDZ3), PSD-95 is a hub for a variety of transient protein-protein interactions. One particularly important role of PSD-95 is as a regulatory connection to neuronal nitric oxide synthase (nNOS). nNOS has been implicated in several neurotoxicity behaviors, prime among these being a result of brain ischemia and stroke. Molecular intervention at the level of PSD-95—namely, to create selective cell-permeable inhibitors that could target each PDZ domain—would provide for both useful research tools to further investigate this pathway. In addition, these ligands could constitute potential therapeutic agents for treating disorders resulting from faulty or unregulated nNOS activity.
Much of our early work explored the fundamental binding properties of PDZ3-mediated molecular interactions. Our group was among the first to apply the powerful technique of isothermal titration calorimetry (ITC) to PDZ domains. Through the integrated use of structural biological data and computer-aided molecular modeling, and more recently chemical library strategies, we have designed and developed high-affinity and selective, cell-permeable ligands to serve as inhibitors of PDZ domain-based protein interactions. These ligands comprise a variety of peptide-based forms, from standard linear to unnatural cyclic and organic-modified acyclic structures. Several of these compounds are now being used in collaborating biology laboratories as molecular probes to decipher the in vivo activities of PDZ domain proteins, especially those that occupy key positions in biochemical pathways that regulate neurobiological processes and cancer.
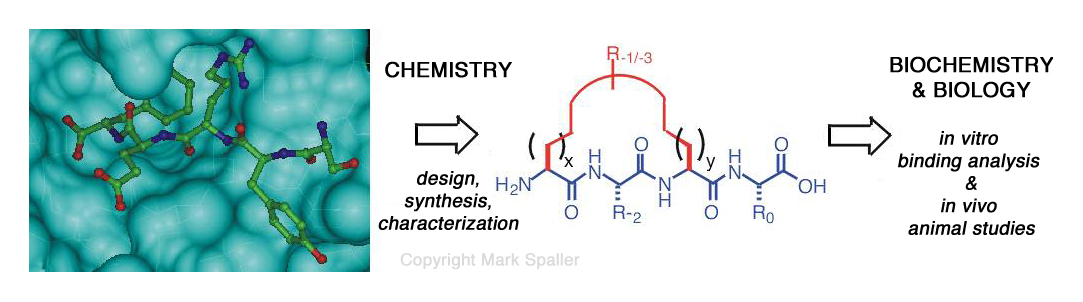
II. Molecular Design & Chemical Synthesis of Bioactive Small Molecules
Peptide-based structures, as employed in our PDZ domain project, are enormously useful as in vitro binding ligands and in vivo probes. 'Non-peptide' organic structures are likewise a useful class, and in fact are of broader utility when pursuing a molecular agent for use as a therapeutic drug. Organic synthesis methods have steadily improved over the decades, contributing regularly to the production of bioactive compounds. But it has been the advent of combinatorial chemical methods and diversity-oriented synthesis design strategies that unleashed the ability to generate more rapidly, and in far greater numbers, organic compounds for use in cellular probe and drug develoment programs. 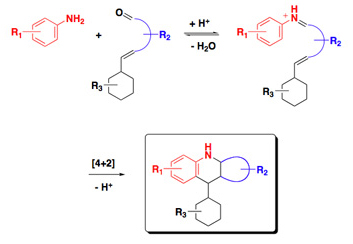 One of the requirements to take adavantage of the technical advances in the area, however, is the availability of 'flexible' multistep synthetic pathways; i.e., those transformations that can accept chemical diverse input, and consistently provide—in good yields—all the corresponding analogs based on those varied starting materials.Such an approach developed in our laboratory incorporated a variant of the intramolecular aza-Diels-Alder cyclization to prepare chemical libraries suitable for cell-based screening. This synthetic route is short, accommodating of many structurally and functionally diverse starting materials, and generates heterocyclic 'drug-like' compounds in good yields.
One of the requirements to take adavantage of the technical advances in the area, however, is the availability of 'flexible' multistep synthetic pathways; i.e., those transformations that can accept chemical diverse input, and consistently provide—in good yields—all the corresponding analogs based on those varied starting materials.Such an approach developed in our laboratory incorporated a variant of the intramolecular aza-Diels-Alder cyclization to prepare chemical libraries suitable for cell-based screening. This synthetic route is short, accommodating of many structurally and functionally diverse starting materials, and generates heterocyclic 'drug-like' compounds in good yields.
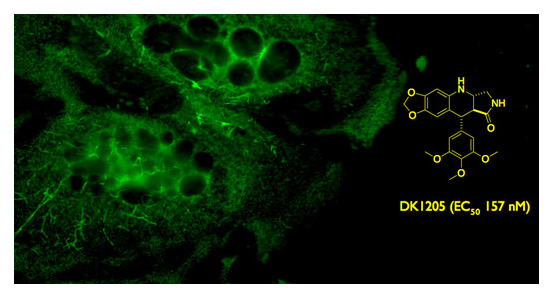 For one project, our objective was to discover new organic structures with anticancer activities. Initiated from a chemistry starting pont, a series of investigations to explore the synthetic scope and limitations of the transformation were required, to allow for efficient production of numerous analogs for testing. After a chemical library was prepared, we entered into the biological phase of the project, in which a series of cell-based assays conducted by collaborating groups to screen for active compounds. Several potent small molecule compounds were discovered, validating our original hypothesis for the design of the analogs. Further studies yielded mechanistic information, implicating microtubules as the site of interaction. Our current work is further adapting and expanding the chemistry we have discovered to prepare novel compounds to target other non-microtubulin proteins implicated in cancer pathways
For one project, our objective was to discover new organic structures with anticancer activities. Initiated from a chemistry starting pont, a series of investigations to explore the synthetic scope and limitations of the transformation were required, to allow for efficient production of numerous analogs for testing. After a chemical library was prepared, we entered into the biological phase of the project, in which a series of cell-based assays conducted by collaborating groups to screen for active compounds. Several potent small molecule compounds were discovered, validating our original hypothesis for the design of the analogs. Further studies yielded mechanistic information, implicating microtubules as the site of interaction. Our current work is further adapting and expanding the chemistry we have discovered to prepare novel compounds to target other non-microtubulin proteins implicated in cancer pathways
For each of the project areas, a separate link provides additional detail:
I. Design & Synthesis of Anticancer Chemical Libraries
II. PDZ Domains & Neurobiology
III. PDZ Domains & Cancer
IV. Protein-Ligand Interactions
