How is gene regulation designed to control the timing of cell processes?
Whether cells are responding to signals in their environment or going about their natural cycle, cell processes depend on the coordinated expression of multiple genes, and this coordination involves much more than merely turning the correct genes on and off. Nowadays we are able to piece together large portions of the complex network connecting the cells genes and other molecular components, but how can we decode the information about the dynamics of this network? Indeed, gene expression is regulated by a variety of mechanisms, and the abundance of binding sites of all sorts imprinted in the DNA highlights the evolutionary importance of creating the patterns of expression that make the cell alive.
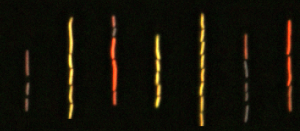
Gene expression is a precise composition of pulses, oscillations and other analog signals. We apply notions of control and circuit design to make sense of biological networks, combining experimental and computational methods. Focusing on antibiotic responses in bacteria, a highly dynamical process, we want to understand how gene regulation is shaped in a dynamical environment. We model the molecular interactions between network components, we use microfluidics to measure the dynamics of gene expression in single cells and we evolve strains under precise conditions using continuous cultures.
How do antibiotic resistance mechanisms adapt to clinical environments?
Antibiotic compounds and their respective resistance mechanisms evolved mostly in the soil, in a bacterial arms race going back perhaps a billion years. Recently, with the clinical use of antibiotics, this arms race has been transported to a very different environment, with different bacterial species involved. While antibiotics in the soil are not thought to reach high concentrations, we treat bacterial infections with periodic administrations of high doses of drugs. What kind of features do resistance mechanisms need to develop to spread through bacterial population in this new environment?
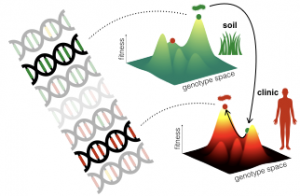
Although antibiotics pose a very strong selective pressure on bacterial populations, this pressure is not constant. Therefore, antibiotic resistance mechanisms are often present in mobile genetic elements, and are often repressed in order to reduce their cost for the cell. We want to find out how the trade-offs between these costs and benefits shape the evolution of antibiotic resistance mechanisms. We analyze the evolutionary history of resistance mechanisms in microbial genome databases, looking for the emergence of new adaptive features. We also reproduce this process in the lab, evolving strains in conditions that mimic the regimens found in different natural environments, such as the soil or the clinic.
How do heterogeneous microbial communities react to changes in their environment?
Bacteria in the natural world exist as complex communities of multiple species that process the nutrients available in their environment together, such as in our microbiome. These different species have different metabolisms, and the diversity of the community is maintained by exchanges of metabolites between these different species. In this context of competition and cooperation, each species must find its niche, fulfilling a particular role in the global behavior of the community, at the risk of being purged in favor of another species that is more fit in that same niche. How then does a complex bacterial community change its composition in response to changes in their environment? What happens when particular species are eliminated when communities are exposed to antibiotics?

A change in the availability of nutrients or stressors, caused by changes in either the environment or community composition, can create new niches and cause a reorganization of the community. We want to predict this reorganization based on knowledge of each species’ individual metabolism and relevant responses. We will propagate defined bacterial communities in continuous cultures under controlled variable environments, measuring each species’ abundance, to develop a predictive model of responses at a community scale.