Current Research Projects
Our laboratory has been engaged in basic and translational research in cholesterol homeostasis. Our goals are to understand cholesterol homeostasis in the CNS and in systemic tissues and to use the knowledge gained as the basis to develop novel therapeutic agents to ameliorate human diseases. We use biochemistry, cell and molecular biology, nanotechnology, medicinal chemistry, as well as mouse models as research tools to pursue our goal.
Cholesterol is a lipid molecule. It is present in the membranes of all mammalian cells. Cells contain different membranes. Each membrane contains certain cholesterol-rich microdomains. These microdomains are associated with specific other lipids as well as specific protein molecules. Proper distributions of these cholesterol-rich microdomains in membranes are required to provide optimal cell growth and maintenance. In many neurodegenerative diseases and in cardiovascular diseases, the individual toxins that cause the specific diseases often disrupt the cholesterol-rich microdomains and cause these microdomains to malfunction.
A favorite research subject in our laboratory is acyl-coenzyme A: cholesterol acyltransferase (ACAT), also named as sterol O-acyltransferase (SOAT). ACAT is an integral membrane protein located in the endoplasmic reticulum. It catalyzes the formation of cholesteryl esters from cholesterol and long-chain fatty acyl coenzyme A. Cholesteryl ester is the storage form of cholesterol, but it cannot substitute the function of cholesterol in membranes. The first gene encoding the enzyme, designated as ACAT1 was identified in our laboratory through an expression cloning approach. We have also purified this protein to homogeneity and characterized it biochemically. ACAT1 is present in many cell types and plays a key role in the cholesterol storage process. In many neurodegenerative diseases, the cholesterol-rich microdomains in various cell types are disrupted. We have shown by a genetic approach that, in mouse models for Alzheimers disease and for Niemann Pick type C disease, blocking cholesterol storage by inactivating ACAT1 can divert the cholesterol storage pool, such that the mobilized cholesterol can repair the disrupted cholesterol-rich microdomains present in the cells. Based on these results, future investigations are directed to develop novel, brain-permeable ACAT inhibitors to ameliorate Alzheimer's disease, Niemann-Pick type C disease, as well as atherosclerosis. We will also use biochemical and biophysical approaches to identify the active sites and regulatory sites in ACAT1. In addition, we will investigate the consequences of inhibiting ACAT in different tissues and cells, including macrophages, neurons, microglial cells, and astrocytes, at the cell and molecular biological level.
Cholesterol Sensing, Trafficking, and Esterification
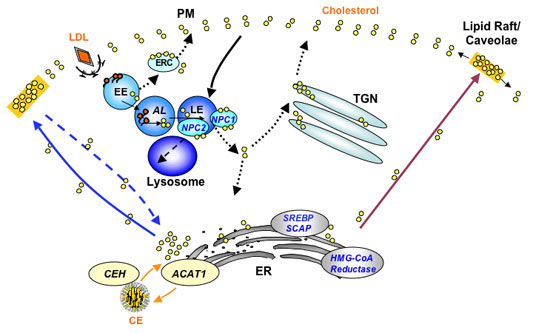
A model for intracellular cholesterol trafficking in a simple mammalian cell. This diagram shows the trafficking/recycling routes of three major cholesterol pools: cholesterol derived from low-density lipoprotein (LDL), cholesterol synthesized de novo in the endoplasmic reticulum (ER), and cholesterol involved in the cholesterol/cholesteryl ester (CE) cycle. The plasma membranes (PMs) contain the highest concentration of cholesterol. The cholesterol-sensing membrane proteins are located in the ER [HMG-CoA reductase (HMGR), SREBP cleavageactivating protein (SCAP), and acyl-coenzyme A:cholesterol acyltransferase 1 (ACAT1)] or in the late endosomes [Niemann-Pick type C1 (NPC1)]. The translocation of cholesterol between various compartments may involve both vesicular and nonvesicular mechanisms. The dotted lines represent cholesterol trafficking steps that are not well documented. Other abbreviations used: AL, acid lipase; CEH, cholesteryl ester hydrolase; EE, early endosome; ERC, endocytic recycling compartment; LE, late endosome; NPC2, Niemann-Pick type C2; SREBP, sterol-regulatory elementbinding protein; TGN, trans-Golgi network. See Review entitled "Cholesterol Sensing,Trafficking, and Esterification" by Chang et al. (2006) for details.
